Filtering¶
Operations looking to remove compounds from a collection are implemented
as Filters in scikit-chem. These are implemented in the
skchem.filters packages:
In [19]:
skchem.filters.__all__
Out[19]:
['ChiralFilter',
'SMARTSFilter',
'PAINSFilter',
'ElementFilter',
'OrganicFilter',
'AtomNumberFilter',
'MassFilter',
'Filter']
They are used very much like Transformers:
In [20]:
of = skchem.filters.OrganicFilter()
In [21]:
benzene = skchem.Mol.from_smiles('c1ccccc1', name='benzene')
ferrocene = skchem.Mol.from_smiles('[cH-]1cccc1.[cH-]1cccc1.[Fe+2]', name='ferrocene')
norbornane = skchem.Mol.from_smiles('C12CCC(C2)CC1', name='norbornane')
dicyclopentadiene = skchem.Mol.from_smiles('C1C=CC2C1C3CC2C=C3')
ms = [benzene, ferrocene, norbornane, dicyclopentadiene]
In [22]:
of.filter(ms)
OrganicFilter: 100% (4 of 4) |#################################################| Elapsed Time: 0:00:00 Time: 0:00:00
Out[22]:
benzene <Mol: c1ccccc1>
norbornane <Mol: C1CC2CCC1C2>
3 <Mol: C1=CC2C3C=CC(C3)C2C1>
Name: structure, dtype: object
Filters essentially use a predicate function to decide whether
to keep or remove instances. The result of this function can be returned
using transform:
In [23]:
of.transform(ms)
OrganicFilter: 100% (4 of 4) |#################################################| Elapsed Time: 0:00:00 Time: 0:00:00
Out[23]:
benzene True
ferrocene False
norbornane True
3 True
dtype: bool
Filters are Transformers¶
As Filters have a transform method, they are themselves
Transformers, that transform a molecule into the result of the
predicate!
In [24]:
issubclass(skchem.filters.Filter, skchem.base.Transformer)
Out[24]:
True
The predicate functions should return None, False or np.nan
for negative results, and anything else for positive results
Creating your own Filter¶
You can create your own filter by passing a predicate function to the
Filter class. For example, perhaps you only wanted compounds to keep
compounds that had a name:
In [25]:
is_named = skchem.filters.Filter(lambda m: m.name is not None)
We carelessly did not set dicyclopentadiene’s name previously, so we want this to get filtered out:
In [26]:
is_named.filter(ms)
Filter: 100% (4 of 4) |########################################################| Elapsed Time: 0:00:00 Time: 0:00:00
Out[26]:
benzene <Mol: c1ccccc1>
ferrocene <Mol: [Fe+2].c1cc[cH-]c1.c1cc[cH-]c1>
norbornane <Mol: C1CC2CCC1C2>
Name: structure, dtype: object
It worked!
Transforming and Filtering¶
A common functionality in cheminformatics is to convert a molecule into something else, and if the conversion fails, to just remove the compound. An example of this is standardization, where one might want to throw away compounds that fail to standardize, or geometry optimization where one might throw away molecules that fail to converge.
This functionality is similar to but crucially different from simply
``filtering``, as filtering returns the original compounds, rather
than the transformed compounds. Instead, there are special
Filters, called TransformFilters, that can perform this task
in a single method call. To give an example of the functionality, we
will use the UFF class:
In [27]:
issubclass(skchem.forcefields.UFF, skchem.filters.base.TransformFilter)
Out[27]:
True
They are instanciated the same way as normal Transformers and
Filters:
In [28]:
uff = skchem.forcefields.UFF()
An example molecule that fails is taken from the NCI DTP Diversity set III:
In [29]:
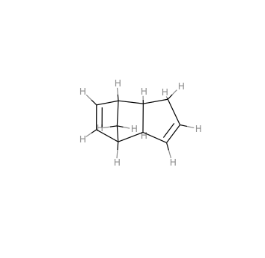
mol_that_fails = skchem.Mol.from_smiles('C[C@H](CCC(=O)O)[C@H]1CC[C@@]2(C)[C@@H]3C(=O)C[C@H]4C(C)(C)[C@@H](O)CC[C@]4(C)[C@H]3C(=O)C[C@]12C',
name='7524')
In [30]:
skchem.vis.draw(mol_that_fails)
Out[30]:
<matplotlib.image.AxesImage at 0x121561eb8>
In [31]:
ms.append(mol_that_fails)
In [32]:
res = uff.filter(ms); res
/Users/rich/projects/scikit-chem/skchem/forcefields/base.py:54: UserWarning: Failed to Embed Molecule 7524
warnings.warn(msg)
UFF: 100% (5 of 5) |###########################################################| Elapsed Time: 0:00:01 Time: 0:00:01
Out[32]:
benzene <Mol: c1ccccc1>
ferrocene <Mol: [Fe+2].c1cc[cH-]c1.c1cc[cH-]c1>
norbornane <Mol: C1CC2CCC1C2>
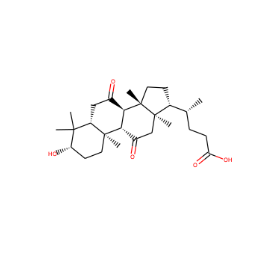
3 <Mol: C1=CC2C3C=CC(C3)C2C1>
Name: structure, dtype: object
Note
filter returns the original molecules, which have not been optimized:
In [33]:
skchem.vis.draw(res.ix[3])
Out[33]:
<matplotlib.image.AxesImage at 0x12174c198>
In [34]:
res = uff.transform_filter(ms); res
/Users/rich/projects/scikit-chem/skchem/forcefields/base.py:54: UserWarning: Failed to Embed Molecule 7524
warnings.warn(msg)
UFF: 100% (5 of 5) |###########################################################| Elapsed Time: 0:00:01 Time: 0:00:01
Out[34]:
benzene <Mol: [H]c1c([H])c([H])c([H])c([H])c1[H]>
ferrocene <Mol: [Fe+2].[H]c1c([H])c([H])[c-]([H])c1[H].[...
norbornane <Mol: [H]C1([H])C([H])([H])C2([H])C([H])([H])C...
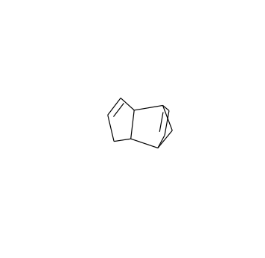
3 <Mol: [H]C1=C([H])C2([H])C3([H])C([H])=C([H])C...
Name: structure, dtype: object
In [35]:
skchem.vis.draw(res.ix[3])
Out[35]:
<matplotlib.image.AxesImage at 0x121925390>